How HIV enters the genome – Researchers identify previously unknown mechanism
This scanning electron microscope image shows a human T cell with numerous yellow-colored HIV-1 virus particles attached to its surface.
Researchers at the German Center for Infection Research (DZIF) at Heidelberg University Hospital have decoded a previously unknown mechanism by which HIV-1 selects its integration targets in the human genome. A research team led by DZIF scientist Dr. Marina Lusic identified RNA:DNA hybrids (R-loops) as molecular signposts for the virus. These findings reveal a key vulnerability in the life cycle of HIV-1. The results, published in the renowned journal Nature Microbiology, provide new therapeutic approaches for specifically controlling HIV reservoirs in the body. This has been one of the biggest obstacles to long-term or curative HIV therapies.
Thanks to antiretroviral therapy, people living with HIV can lead almost normal lives. Antiretroviral drugs prevent the virus from multiplying, but must be taken daily for life. However, any interruption in treatment—due to limited access, supply disruptions, or adherence challenges—can result in rapid viral rebound and, more worryingly, the emergence of drug-resistant HIV variants.
The HIV virus primarily infects cells of the immune system, anchoring its genetic material in T cells in particular. Once integrated, these viral sequences create lifelong reservoir of infection. The HIV-1 integrase enzyme is responsible for inserting the virus into the host genome, forcing cells to produce new viruses and enabling the ongoing infection process. "Until now, it has been not entirely clear how HIV-1 integrase selects its integration targets in the genome. A deeper understanding of this process is crucial for developing new treatment strategies and tackling the persistent viral reservoirs that cannot be eliminated by existing therapies," says Dr. Marina Lusic, DZIF scientist at the Center for Integrative Infectious Disease Research (CIID) at Heidelberg University Hospital, who led the study.
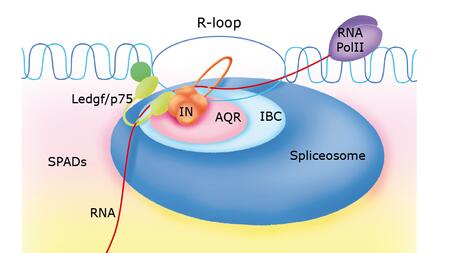
RNA:DNA hybrids as signposts for virus integration
The research team was able to prove that HIV-1 does not randomly invade the genome, but uses specific signposts: so-called RNA:DNA hybrids or “R-loops”, which mainly occur in non-coding regions of active genes. The researchers mapped these structures in human immune cells and demonstrated that the viral integrase docks precisely at these locations. “The virus follows these structures like signposts on a map and thus finds the appropriate integration sites,” explains Dr. Carlotta Penzo, senior postdoctoral researcher in Dr. Marina Lusic's team and first author of the study. “Another important result of our investigation is that a specific cellular partner, the enzyme Aquarius, helps the virus in R-loop recognition, enabling HIV-1 insertion into RNA:DNA hybrids.”
The splicing enzyme RNA helicase Aquarius (AQR) plays a key role in this process. It acts as a kind of door opener, binding to HIV-1 integrase and promoting integration by unwinding the R-loops. “Our results show that removing AQR significantly decreases the integration rate. The remaining integration events shift to R-loop-poor regions—clear evidence of the association between viral integration and AQR activity on R-loops,” says Penzo.
“This discovery opens a new avenue for HIV intervention. If we can disrupt the virus’s ability to use host RNA structures for integration, we may be able to limit or redirect where HIV hides and ultimately reduce or eliminate the need for lifelong therapy,” says Dr. Marina Lusic. “These findings are particularly significant in light of the increasing global instability in HIV care. In many regions, the continuous provision of antiretroviral therapies is not guaranteed—with the result that interruptions significantly increase the risk of treatment failure and the spread of resistant virus variants.”
The results reveal previously unknown targets for combating HIV. In the long term, the identified R-loop/Aquarius mechanism could help to specifically target HIV reservoirs in the body that existing therapies cannot eliminate—and thus point the way to new, effective, and potentially curative forms of treatment.
Funding and international collaboration
This study was supported by the German Center for Infection Research (Deutsches Zentrum für Infektionsforschung, DZIF) and by the German Research Foundation (DFG) through the Special Collaborative Programme SFB 1129. It was conducted through a multidisciplinary collaboration led by the group of Dr. Marina Lusic, in partnership with colleagues from the Center for Integrative Infectious Disease Research (CIID) Heidelberg, including Prof. Oliver Fackler and Prof. Hans-Georg Kräusslich. Furthermore, the study was made possible by close pan-European collaboration, with contributions from bioinformatics, structural biology and retrovirology experts at research institutions in Zagreb, Padua, London and Bordeaux.